Motifs detected by DREME on the MeRIP-seq datasets
5 (218) · € 13.00 · En Stock
Download scientific diagram | Motifs detected by DREME on the MeRIP-seq datasets from publication: A novel algorithm for calling mRNA m 6 A peaks by modeling biological variances in MeRIP-seq data | Motivation: N⁶-methyl-adenosine (m⁶A) is the most prevalent mRNA methylation but precise prediction of its mRNA location is important for understanding its function. A recent sequencing technology, known as Methylated RNA Immunoprecipitation Sequencing technology (MeRIP-seq), | mRNA, RNA Sequence Analysis and Base Sequence | ResearchGate, the professional network for scientists.

A hierarchical model for clustering m6A methylation peaks in MeRIP-seq data, BMC Genomics
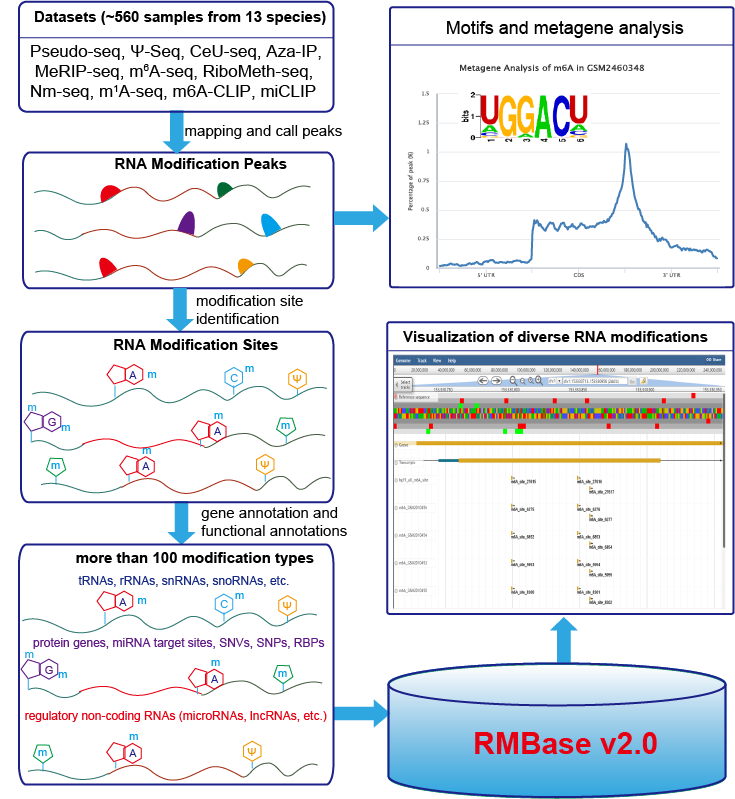
RNA Modification Database, RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data (Pseudo-seq, Ψ-seq, CeU- seq, Aza-IP, MeRIP-seq, m6A-seq, RiboMeth-seq, m1A-seq).
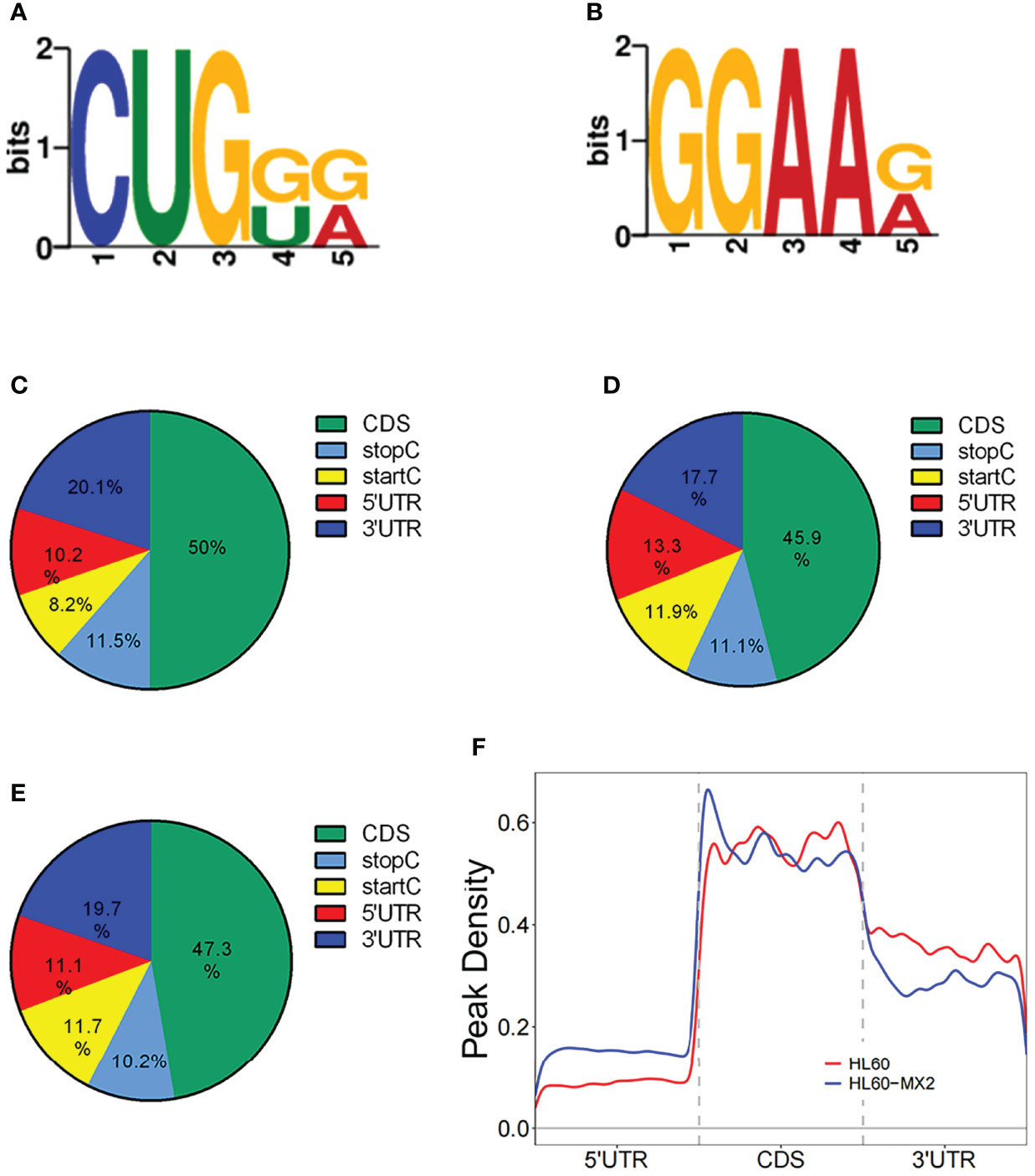
Frontiers Transcriptome Profiling of N7-Methylguanosine Modification of Messenger RNA in Drug-Resistant Acute Myeloid Leukemia

Two most significant motifs found by the MEME, DREME and AME algorithms

DENA: training an authentic neural network model using Nanopore sequencing data of Arabidopsis transcripts for detection and quantification of N6-methyladenosine on RNA

A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. - Abstract - Europe PMC

A hierarchical model for clustering m6A methylation peaks in MeRIP-seq data, BMC Genomics

Limits in the detection of m6A changes using MeRIP/m6A-seq

A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package - ScienceDirect

Motifs detected by DREME on the MeRIP-seq datasets
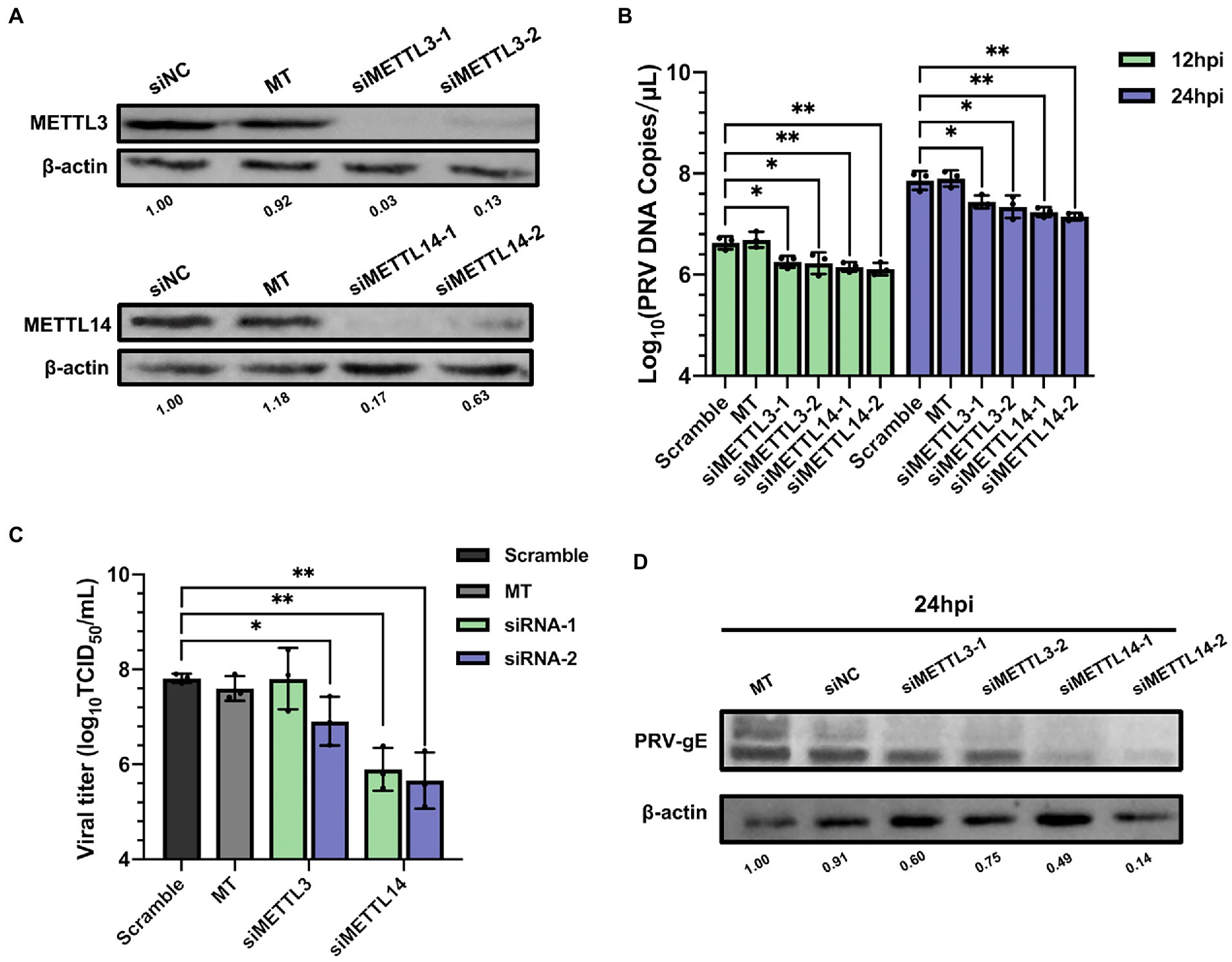
Frontiers Pseudorabies virus exploits N6-methyladenosine modification to promote viral replication
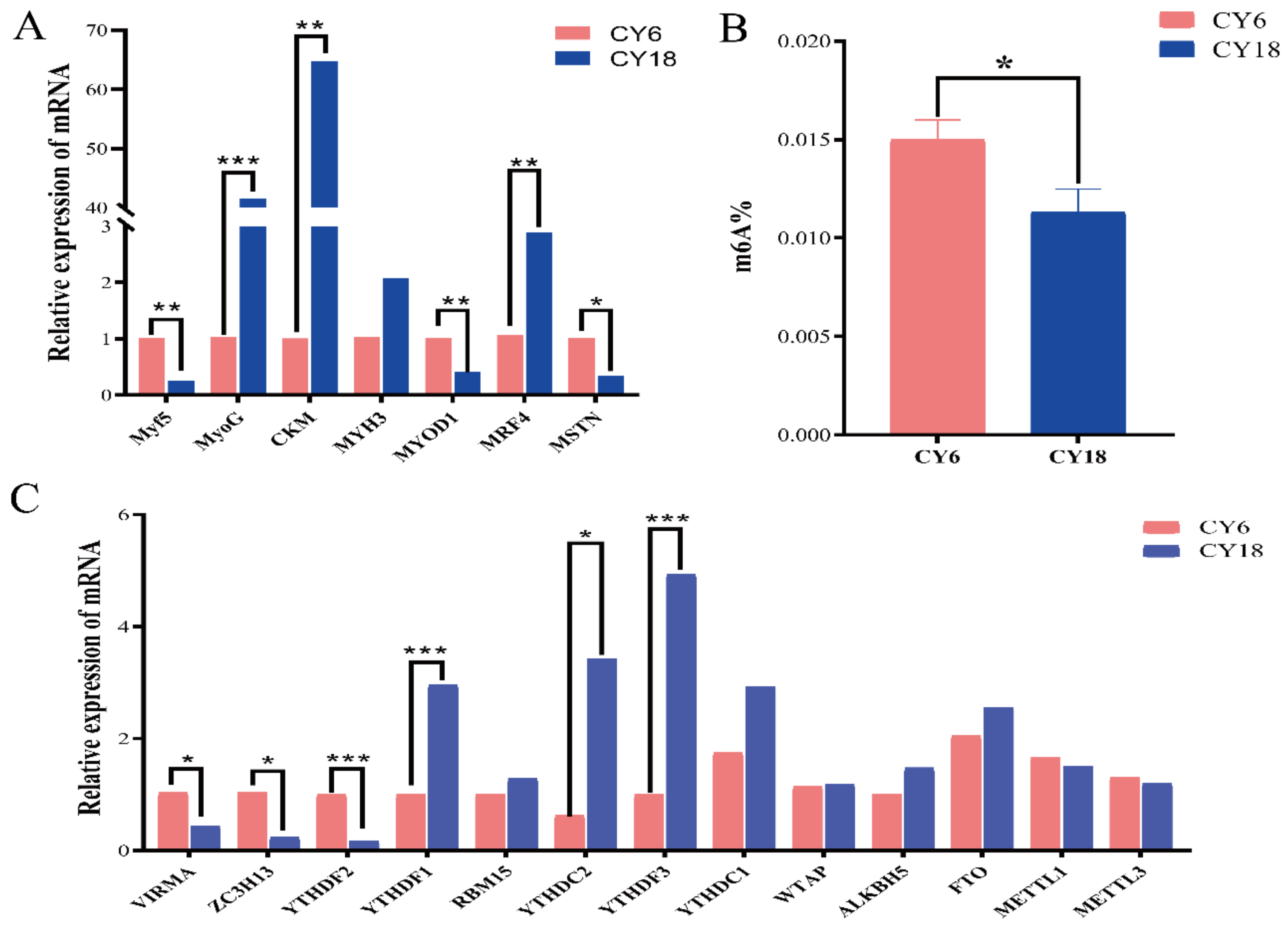
Cells, Free Full-Text
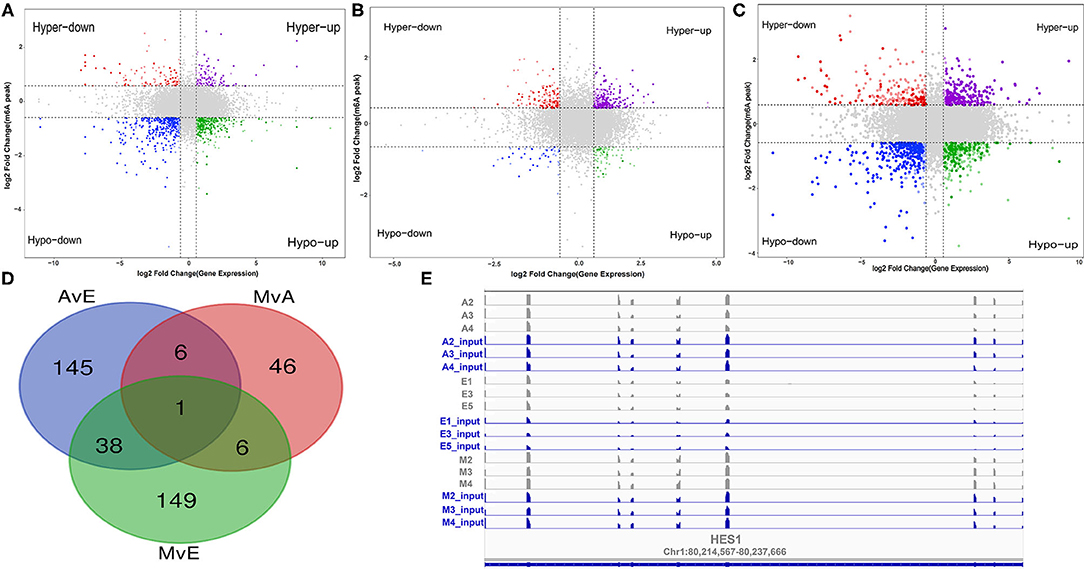
Frontiers Regulatory Role of N6-Methyladenosine in Longissimus Dorsi Development in Yak

A hierarchical model for clustering m6A methylation peaks in MeRIP-seq data, BMC Genomics

Motif and regional analysis of m7G methylation. (A) Motif with